 W
WCdc6, or cell division cycle 6, is a protein in eukaryotic cells. It is mainly studied in the budding yeast Saccharomyces cerevisiae. It is an essential regulator of DNA replication and plays important roles in the activation and maintenance of the checkpoint mechanisms in the cell cycle that coordinate S phase and mitosis. It is part of the pre-replicative complex (pre-RC) and is required for loading minichromosome maintenance (MCM) proteins onto the DNA, an essential step in the initiation of DNA synthesis. In addition, it is a member of the family of AAA+ ATPases and highly related to ORC1; both are the same protein in archaea.
 W
WThe centromere is the specialized DNA sequence of a chromosome that links a pair of sister chromatids. During mitosis, spindle fibers attach to the centromere via the kinetochore. Centromeres were first thought to be genetic loci that direct the behavior of chromosomes.
 W
WChromatin assembly factor-1 (CAF-1) is a protein complex — including Chaf1a (p150), Chaf1b (p60), and p50 subunits — that assembles histone tetramers onto replicating DNA. CAF-1 functions as a histone chaperone that mediates the first step in nucleosome formation by tetramerizing and depositing newly synthesized histone H3/H4 onto DNA rapidly behind replication forks. Several studies have shown that the interaction between CAF-1 and PCNA, which stabilizes CAF-1 at replication forks, is important for CAF-1's role in nucleosome assembly
 W
WIn cell biology, eukaryotes possess a regulatory system that ensures that DNA replication occurs only once per cell cycle.
 W
WA DNA clamp, also known as a sliding clamp or β-clamp, is a protein complex that serves as a processivity-promoting factor in DNA replication. As a critical component of the DNA polymerase III holoenzyme, the clamp protein binds DNA polymerase and prevents this enzyme from dissociating from the template DNA strand. The clamp-polymerase protein–protein interactions are stronger and more specific than the direct interactions between the polymerase and the template DNA strand; because one of the rate-limiting steps in the DNA synthesis reaction is the association of the polymerase with the DNA template, the presence of the sliding clamp dramatically increases the number of nucleotides that the polymerase can add to the growing strand per association event. The presence of the DNA clamp can increase the rate of DNA synthesis up to 1,000-fold compared with a nonprocessive polymerase.
 W
WDNA ligase is a specific type of enzyme, a ligase, that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living organisms, but some forms may specifically repair double-strand breaks. Single-strand breaks are repaired by DNA ligase using the complementary strand of the double helix as a template, with DNA ligase creating the final phosphodiester bond to fully repair the DNA.
 W
WA DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reactiondeoxynucleoside triphosphate + DNAn ⇌ pyrophosphate + DNAn+1.
 W
WDNA polymerase alpha also known as Pol α is an enzyme complex found in eukaryotes that is involved in initiation of DNA replication. The DNA polymerase alpha complex consists of 4 subunits: POLA1, POLA2, PRIM1, and PRIM2.
 W
WDNA polymerase delta (DNA Pol δ) is an enzyme complex found in eukaryotes that is involved in DNA replication and repair. The DNA polymerase delta complex consists of 4 subunits: POLD1, POLD2, POLD3, and POLD4. DNA Pol δ is an enzyme used for both leading and lagging strand synthesis. It exhibits increased processivity when interacting with the proliferating cell nuclear antigen (PCNA). As well, the multisubunit protein replication factor C, through its role as the clamp loader for PCNA is important for DNA Pol δ function.
 W
WDNA polymerase eta, is a protein that in humans is encoded by the POLH gene.
 W
WDNA polymerase I is an enzyme that participates in the process of prokaryotic DNA replication. Discovered by Arthur Kornberg in 1956, it was the first known DNA polymerase. It was initially characterized in E. coli and is ubiquitous in prokaryotes. In E. coli and many other bacteria, the gene that encodes Pol I is known as polA. The E. coli Pol I enzyme is composed of 928 amino acids, and is an example of a processive enzyme — it can sequentially catalyze multiple polymerisation steps without releasing the single-stranded template. The physiological function of Pol I is mainly to support repair of damaged DNA, but it also contributes to connecting Okazaki fragments by deleting RNA primers and replacing the ribonucleotides with DNA.
 W
WDNA polymerase II is a prokaryotic DNA-Dependent DNA polymerase encoded by the PolB gene.
 W
WDNA polymerase III holoenzyme is the primary enzyme complex involved in prokaryotic DNA replication. It was discovered by Thomas Kornberg and Malcolm Gefter in 1970. The complex has high processivity and, specifically referring to the replication of the E.coli genome, works in conjunction with four other DNA polymerases. Being the primary holoenzyme involved in replication activity, the DNA Pol III holoenzyme also has proofreading capabilities that corrects replication mistakes by means of exonuclease activity reading 3'→5' and synthesizing 5'→3'. DNA Pol III is a component of the replisome, which is located at the replication fork.
 W
WIn molecular biology, the δ (delta) subunit of DNA polymerase III is encoded by the holA gene in E. coli and other bacteria. Along with the γ, δ', χ, and ψ subunits that make up the core polymerase, and the β accessory proteins, the δ subunit is responsible for the high speed and processivity of polIII.
 W
WPolymerase nu is a protein in humans that is encoded by the POLN gene. It is a family A DNA polymerase, considered to be the least effective of the polymerase enzymes. However, DNA polymerase nu plays an active role in homology repair during cellular responses to crosslinks, fulfilling its role in a complex with helicase.
 W
WIn molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential.
 W
WCDT1 is a protein that in humans is encoded by the CDT1 gene. It is a licensing factor that functions to limit DNA from replicating more than once per cell cycle.
 W
WDNA replication stress refers to the state of a cell whose genome is exposed to various stresses. The events that contribute to replication stress occur during DNA replication, and can result in a stalled replication fork.
 W
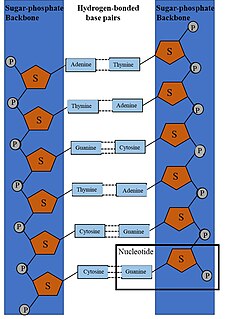
WDNA synthesis is the natural or artificial creation of deoxyribonucleic acid (DNA) molecules. DNA is a macromolecule made up of nucleotide units, which are linked by covalent bonds and hydrogen bonds, in a repeating structure. DNA synthesis occurs when these nucleotide units are joined together to form DNA; this can occur artificially or naturally. Nucleotide units are made up of a nitrogenous base, pentose sugar (deoxyribose) and phosphate group. Each unit is joined when a covalent bond forms between its phosphate group and the pentose sugar of the next nucleotide, forming a sugar-phosphate backbone. DNA is a complementary, double stranded structure as specific base pairing occurs naturally when hydrogen bonds form between the nucleotide bases.
 W
WDNA-binding proteins are proteins that have DNA-binding domains and thus have a specific or general affinity for single- or double-stranded DNA. Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, because it exposes more functional groups that identify a base pair. However, there are some known minor groove DNA-binding ligands such as netropsin, distamycin, Hoechst 33258, pentamidine, DAPI and others.
 W
WDnaA is a protein that activates initiation of DNA replication in bacteria. It is a replication initiation factor which promotes the unwinding of DNA at oriC. The onset of the initiation phase of DNA replication is determined by the concentration of DnaA. DnaA accumulates during growth and then triggers the initiation of replication. Replication begins with active DnaA binding to 9-mer (9-bp) repeats upstream of oriC. Binding of DnaA leads to strand separation at the 13-mer repeats. This binding causes the DNA to loop in preparation for melting open by the helicase DnaB.
 W
WDnaB helicase is an enzyme in bacteria which opens the replication fork during DNA replication. Although the mechanism by which DnaB both couples ATP hydrolysis to translocation along DNA and denatures the duplex is unknown, a change in the quaternary structure of the protein involving dimerisation of the N-terminal domain has been observed and may occur during the enzymatic cycle. Initially when DnaB binds to dnaA, it is associated with dnaC, a negative regulator. After DnaC dissociates, DnaB binds dnaG.
 W
WdnaN is the gene that codes for the DNA clamp of DNA polymerase III in prokaryotes. The β clamp physically locks Pol III onto a DNA strand during replication to help increase its processivity. The eukaryotic equivalent to the β clamp is PCNA.
 W
WEukaryotic DNA replication is a conserved mechanism that restricts DNA replication to once per cell cycle. Eukaryotic DNA replication of chromosomal DNA is central for the duplication of a cell and is necessary for the maintenance of the eukaryotic genome.
 W
WHelicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genes. They are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two annealed nucleic acid strands such as DNA and RNA, using energy from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases. The human genome codes for 95 non-redundant helicases: 64 RNA helicases and 31 DNA helicases. Many cellular processes, such as DNA replication, transcription, translation, recombination, DNA repair, and ribosome biogenesis involve the separation of nucleic acid strands that necessitates the use of helicases.
 W
WHelicase, POLQ-like, also known as hel308 and Holliday junction migration protein, encoded by the gene HEL308, is a DNA helicase found in humans, archea and many other organisms. Its principal function is to allow DNA replication to continue past DNA forks.
 W
WIn E. coli and other bacteria, holC is a gene that encodes the chi subunit of DNA polymerase III.
 W
WIn E. coli and other bacteria, holE is a gene that encodes the theta subunit of DNA polymerase III.
 W
WThe Klenow fragment is a large protein fragment produced when DNA polymerase I from E. coli is enzymatically cleaved by the protease subtilisin. First reported in 1970, it retains the 5' → 3' polymerase activity and the 3’ → 5’ exonuclease activity for removal of precoding nucleotides and proofreading, but loses its 5' → 3' exonuclease activity.
 W
WMicrocosm: E. coli and the New Science of Life is a 2008 book by science writer Carl Zimmer. The book presents an overview of genetics research and genetic engineering by telling the story about the Escherichia coli species of bacteria which is omnipresent in the mammalian gastrointestinal tract. The title Microcosm refers to the notion that insights derived from the study of a relatively simple, single-celled organism like E. coli play in describing the fundamental features of all terrestrial life, including humans.
 W
WThe minichromosome maintenance protein complex (MCM) is a DNA helicase essential for genomic DNA replication. Eukaryotic MCM consists of six gene products, Mcm2–7, which form a heterohexamer. As a critical protein for cell division, MCM is also the target of various checkpoint pathways, such as the S-phase entry and S-phase arrest checkpoints. Both the loading and activation of MCM helicase are strictly regulated and are coupled to cell growth cycles. Deregulation of MCM function has been linked to genomic instability and a variety of carcinomas.
 W
WThe monocentric chromosome is a chromosome that has only one centromere in a chromosome and forms a narrow constriction.
 W
WMolecular biologists use several shorthand terms when referring to nucleic acid molecules, such as DNA and RNA, collectively referred to as nucleic acid nomenclature.
 W
WOkazaki fragments are short sequences of DNA nucleotides which are synthesized discontinuously and later linked together by the enzyme DNA ligase to create the lagging strand during DNA replication. They were discovered in the 1960s by the Japanese molecular biologists Reiji and Tsuneko Okazaki, along with the help of some of their colleagues.
 W
WThe origin of replication is a particular sequence in a genome at which replication is initiated. Propagation of the genetic material between generations requires timely and accurate duplication of DNA by semiconservative replication prior to cell division to ensure each daughter cell receives the full complement of chromosomes. This can either involve the replication of DNA in living organisms such as prokaryotes and eukaryotes, or that of DNA or RNA in viruses, such as double-stranded RNA viruses. Synthesis of daughter strands starts at discrete sites, termed replication origins, and proceeds in a bidirectional manner until all genomic DNA is replicated. Despite the fundamental nature of these events, organisms have evolved surprisingly divergent strategies that control replication onset. Although the specific replication origin organization structure and recognition varies from species to species, some common characteristics are shared.
 W
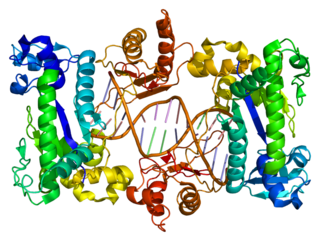
WDNA polymerase subunit gamma is an enzyme that in humans is encoded by the POLG gene. Mitochondrial DNA polymerase is heterotrimeric, consisting of a homodimer of accessory subunits plus a catalytic subunit. The protein encoded by this gene is the catalytic subunit of mitochondrial DNA polymerase. Defects in this gene are a cause of progressive external ophthalmoplegia with mitochondrial DNA deletions 1 (PEOA1), sensory ataxic neuropathy dysarthria and ophthalmoparesis (SANDO), Alpers-Huttenlocher syndrome (AHS), and mitochondrial neurogastrointestinal encephalopathy syndrome (MNGIE).
 W
WDNA polymerase iota is an enzyme that in humans is encoded by the POLI gene. It is found in higher eukaryotes, and is believed to have arisen from a gene duplication from Pol η. Pol ι, is a Y family polymerase that is involved in translesion synthesis. It can bypass 6-4 pyrimidine adducts and abasic sites and has a high frequency of wrong base incorporation. Like many other Y family polymerases Pol ι, has low processivity, a large DNA binding pocket and doesn't undergo conformational changes when DNA binds. These attributes are what allow Pol ι to carry out its task as a translesion polymerase. Pol ι only uses Hoogsteen base pairing, during DNA synthesis, it will add adenine opposite to thymine in the syn conformation and can add both cytosine and thymine in the anti conformation across guanine, which it flips to the syn conformation.
 W
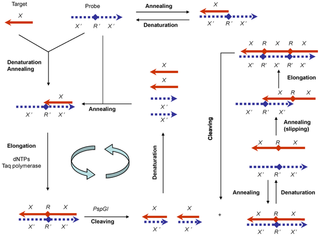
WPolymerase-endonuclease amplification reaction (PEAR) is a DNA amplification technology for the amplification of oligonucleotides. A target oligonucleotide and a tandem repeated antisense probe are subjected to repeated cycles of denaturing, annealing, elongation and cleaving, in which thermostable DNA polymerase elongation and strand slipping generate duplex tandem repeats, and thermostable endonuclease (PspGI) cleavage releases monomeric duplex oligonucleotides.
 W
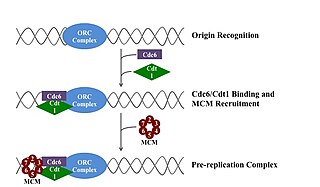
WA pre-replication complex (pre-RC) is a protein complex that forms at the origin of replication during the initiation step of DNA replication. Formation of the pre-RC is required for DNA replication to occur. Complete and faithful replication of the genome ensures that each daughter cell will carry the same genetic information as the parent cell. Accordingly, formation of the pre-RC is a very important part of the cell cycle.
 W
WA primer is a short single-stranded nucleic acid used by all living organisms in the initiation of DNA synthesis. DNA polymerase enzymes are only capable of adding nucleotides to the 3’-end of an existing nucleic acid, requiring a primer be bound to the template before DNA polymerase can begin a complementary strand. Living organisms use solely RNA primers, while laboratory techniques in biochemistry and molecular biology that require in vitro DNA synthesis usually use DNA primers, since they are more temperature stable.
 W
WPrimPol is a protein encoded by the PRIMPOL gene in humans. PrimPol is a eukaryotic protein with both DNA polymerase and DNA Primase activities involved in translesion DNA synthesis. It is the first eukaryotic protein to be identified with priming activity using deoxyribonucleotides. It is also the first protein identified in the mitochondria to have translesion DNA synthesis activities.
 W
WProkaryotic DNA Replication is the process by which a prokaryote duplicates its DNA into another copy that is passed on to daughter cells. Although it is often studied in the model organism E. coli, other bacteria show many similarities. Replication is bi-directional and originates at a single origin of replication (OriC). It consists of three steps: Initiation, elongation, and termination.
 W
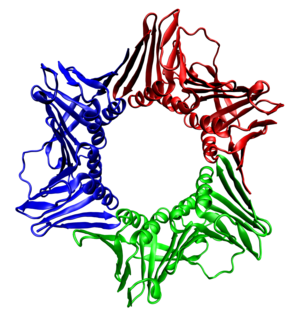
WProliferating cell nuclear antigen (PCNA) is a DNA clamp that acts as a processivity factor for DNA polymerase δ in eukaryotic cells and is essential for replication. PCNA is a homotrimer and achieves its processivity by encircling the DNA, where it acts as a scaffold to recruit proteins involved in DNA replication, DNA repair, chromatin remodeling and epigenetics.
 W
WSlipped strand mispairing (SSM),, is a mutation process which occurs during DNA replication. It involves denaturation and displacement of the DNA strands, resulting in mispairing of the complementary bases. Slipped strand mispairing is one explanation for the origin and evolution of repetitive DNA sequences.
 W
WTus, also known as terminus utilization substance, is a protein that binds to terminator sequences and acts as a counter-helicase when it comes in contact with an advancing helicase. The bound Tus protein effectively halts DNA polymerase movement. Tus helps end DNA replication in prokaryotes.
 W
WReplication timing refers to the order in which segments of DNA along the length of a chromosome are duplicated.
 W
WThe replisome is a complex molecular machine that carries out replication of DNA. The replisome first unwinds double stranded DNA into two single strands. For each of the resulting single strands, a new complementary sequence of DNA is synthesized. The net result is formation of two new double stranded DNA sequences that are exact copies of the original double stranded DNA sequence.
 W
WRepressor LexA or LexA is a transcriptional repressor that represses SOS response genes coding primarily for error-prone DNA polymerases, DNA repair enzymes and cell division inhibitors. LexA forms de facto a two-component regulatory system with RecA, which senses DNA damage at stalled replication forks, forming monofilaments and acquiring an active conformation capable of binding to LexA and causing LexA to cleave itself, in a process called autoproteolysis.
 W
WThe retinoblastoma protein is a tumor suppressor protein that is dysfunctional in several major cancers. One function of pRb is to prevent excessive cell growth by inhibiting cell cycle progression until a cell is ready to divide. When the cell is ready to divide, pRb is phosphorylated, inactivating it, and the cell cycle is allowed to progress. It is also a recruiter of several chromatin remodeling enzymes such as methylases and acetylases.
 W
WRolling circle replication (RCR) is a process of unidirectional nucleic acid replication that can rapidly synthesize multiple copies of circular molecules of DNA or RNA, such as plasmids, the genomes of bacteriophages, and the circular RNA genome of viroids. Some eukaryotic viruses also replicate their DNA or RNA via the rolling circle mechanism.
 W
WRRM3 is a gene that encodes a 5′-to-3′ DNA helicase known affect multiple cellular replication and repair processes and is most commonly studied in Saccharomyces cerevisiae. RRM3 formally stands for Ribosomal DNArecombination mutation 3. The gene codes for nuclear protein Rrm3p, which is 723 amino acids in length, and is part of a Pif1p DNA helicase sub-family that is conserved from yeasts to humans. RRM3 and its encoded protein have been shown to be vital for cellular replication, specifically associating with replication forks genome-wide. RRM3 is located on chromosome 8 in yeast cells and codes for 723 amino acids producing a protein that weighs 81,581 Da.
 W
WThe Selfish Gene is a 1976 book on evolution by the ethologist Richard Dawkins, in which the author builds upon the principal theory of George C. Williams's Adaptation and Natural Selection (1966). Dawkins uses the term "selfish gene" as a way of expressing the gene-centred view of evolution, popularising ideas developed during the 1960s by W. D. Hamilton and others. From the gene-centred view, it follows that the more two individuals are genetically related, the more sense it makes for them to behave cooperatively with each other.
 W
WSingle-stranded binding proteins (SSBs) are a class of proteins that have been identified in both viruses and organisms from bacteria to humans.
 W
WSlipped strand mispairing (SSM),, is a mutation process which occurs during DNA replication. It involves denaturation and displacement of the DNA strands, resulting in mispairing of the complementary bases. Slipped strand mispairing is one explanation for the origin and evolution of repetitive DNA sequences.
 W
WT7 DNA polymerase is an enzyme used during the DNA replication of the T7 bacteriophage. During this process, the DNA polymerase “reads” existing DNA strands and creates two new strands that match the existing ones. The T7 DNA polymerase requires a host factor, E. coli thioredoxin, in order to carry out its function. This helps stabilize the binding of the necessary protein to the primer-template to improve processivity by more than 100-fold, which is a feature unique to this enzyme. It is a member of the Family A DNA polymerases, which include E. coli DNA polymerase I and Taq DNA polymerase.
 W
WTaq polymerase is a thermostable DNA polymerase I named after the thermophilic eubacterial microorganism Thermus aquaticus, from which it was originally isolated by Chien et al. in 1976. Its name is often abbreviated to Taq or Taq pol. It is frequently used in the polymerase chain reaction (PCR), a method for greatly amplifying the quantity of short segments of DNA.
 W
WTelomerase, also called terminal transferase, is a ribonucleoprotein that adds a species-dependent telomere repeat sequence to the 3' end of telomeres. A telomere is a region of repetitive sequences at each end of the chromosomes of most eukaryotes. Telomeres protect the end of the chromosome from DNA damage or from fusion with neighbouring chromosomes. The fruit fly Drosophila melanogaster lacks telomerase, but instead uses retrotransposons to maintain telomeres.
 W
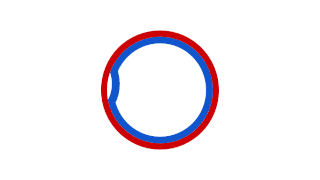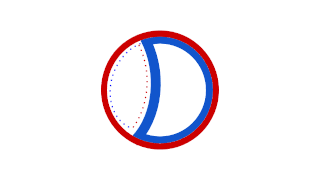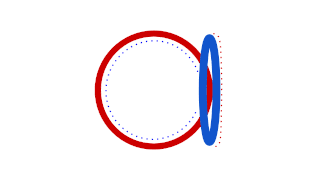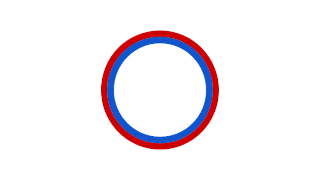
WA theta structure is an intermediate structure formed during the replication of a circular DNA molecule. Two replication forks can proceed independently around the DNA ring and when viewed from above the structure resembles the Greek letter "theta" (θ). Originally discovered by John Cairns, it led to the understanding that bidirectional DNA replication could take place. Proof of the bidirectional nature came from providing replicating cells with a pulse of tritiated thymidine, quenching rapidly and then autoradiographing. Results showed that the radioactive thymidine was incorporated into both forks of the theta structure, not just one, indicating synthesis at both forks in opposite directions around the loop.
 W
WXPB is an ATP-dependent DNA helicase in humans that is a part of the TFIIH transcription factor complex.