 W
WDNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in as many as 1 million individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages. This can eventually lead to malignant tumors, or cancer as per the two hit hypothesis.
 W
WPoly(ADP-ribose) polymerase family member 14 is a protein that, in humans, is encoded by the PARP14 gene.
 W
WAlkD is an enzyme belonging to a family of DNA glycosylases that are involved in DNA repair. It was discovered by a team of Norwegian biologists from Oslo in 2006. It was isolated from a soil-dwelling Gram-positive bacteria Bacillus cereus, along with another enzyme AlkC. AlkC and AlkD are most probably derived from the same protein as indicated by their close resemblance. They are also found in other prokaryotes. Among eukaryotes, they are found only in the single-celled species only, such as Entamoeba histolytica and Dictyostelium discoideum. The enzyme specifically targets 7mG (methyl-guanine) in the DNA, and is, therefore, unique among DNA glycosylases. It can also act on other methylpurines with less affinity. It indicates that the enzyme is specific for locating and cutting (excision) of chemically modified bases from DNA, exactly at 7mG, whenever there are errors in replication. It accelerates the rate of 7mG hydrolysis 100-fold over the spontaneous depurination. Thus, it protects the genome from harmful changes induced by chemical and environmental agents. Its crystal structure was described in 2008. It is the first HEAT repeat protein identified to interact with nucleic acids or to contain enzymatic activity.
 W
WIn biochemistry and molecular genetics, an AP site, also known as an abasic site, is a location in DNA that has neither a purine nor a pyrimidine base, either spontaneously or due to DNA damage. It has been estimated that under physiological conditions 10,000 apurinic sites and 500 apyrimidinic may be generated in a cell daily.
 W
WBase excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from the genome. The related nucleotide excision repair pathway repairs bulky helix-distorting lesions. BER is important for removing damaged bases that could otherwise cause mutations by mispairing or lead to breaks in DNA during replication. BER is initiated by DNA glycosylases, which recognize and remove specific damaged or inappropriate bases, forming AP sites. These are then cleaved by an AP endonuclease. The resulting single-strand break can then be processed by either short-patch or long-patch BER.
 W
WBioSentinel is a planned low-cost CubeSat spacecraft on a space biology mission that will use budding yeast to detect, measure, and compare the impact of deep space radiation on DNA repair over long time beyond low-Earth orbit.
 W
WCell cycle regulator of non-homologous end joining is a protein that in humans is encoded by the CYREN gene.
 W
WDeinococcus radiodurans is an extremophilic bacterium and one of the most radiation-resistant organisms known. It can survive cold, dehydration, vacuum, and acid, and therefore, is known as a polyextremophile and it has been listed as the world's toughest known bacterium in The Guinness Book Of World Records.
 W
WDNA mismatch repair (MMR) is a system for recognizing and repairing erroneous insertion, deletion, and mis-incorporation of bases that can arise during DNA replication and recombination, as well as repairing some forms of DNA damage.
 W
WDNA polymerase beta, also known as POLB, is an enzyme present in eukaryotes. In humans, it is encoded by the POLB gene.
 W
WDNA polymerase delta (DNA Pol δ) is an enzyme complex found in eukaryotes that is involved in DNA replication and repair. The DNA polymerase delta complex consists of 4 subunits: POLD1, POLD2, POLD3, and POLD4. DNA Pol δ is an enzyme used for both leading and lagging strand synthesis. It exhibits increased processivity when interacting with the proliferating cell nuclear antigen (PCNA). As well, the multisubunit protein replication factor C, through its role as the clamp loader for PCNA is important for DNA Pol δ function.
 W
WDNA polymerase lambda, also known as Pol λ, is an enzyme found in all eukaryotes. In humans, it is encoded by the POLL gene.
 W
WDNA polymerase mu is a polymerase enzyme found in eukaryotes. In humans, this protein is encoded by the POLM gene.
 W
WDNA Repair and Mutagenesis is a college-level textbook about DNA repair and mutagenesis written by Errol Friedberg, Graham Walker, Wolfram Siede, Richard D. Wood, and Roger Schultz. In its second edition as of 2009, DNA Repair and Mutagenesis contains over 1,000 pages, 10,000 references and 700 illustrations and has been described as "the most comprehensive book available in [the] field."
 W
WDNA repair protein XRCC4 also known as X-ray repair cross-complementing protein 4 or XRCC4 is a protein that in humans is encoded by the XRCC4 gene. In addition to humans, the XRCC4 protein is also expressed in many other metazoans, fungi and in plants. The X-ray repair cross-complementing protein 4 is one of several core proteins involved in the non-homologous end joining (NHEJ) pathway to repair DNA double strand breaks (DSBs).
 W
WThe G2-M DNA damage checkpoint is an important cell cycle checkpoint in eukaryotic organisms that ensures that cells don't initiate mitosis until damaged or incompletely replicated DNA is sufficiently repaired. Cells which have a defective G2-M checkpoint, if they enter M phase before repairing their DNA, it leads to apoptosis or death after cell division. The defining biochemical feature of this checkpoint is the activation of M-phase cyclin-CDK complexes, which phosphorylate proteins that promote spindle assembly and bring the cell to metaphase.
 W
WH2A histone family member X is a type of histone protein from the H2A family encoded by the H2AFX gene. An important phosphorylated form is γH2AX (140S), which forms when double-strand breaks appear.
 W
WHelicase, POLQ-like, also known as hel308 and Holliday junction migration protein, encoded by the gene HEL308, is a DNA helicase found in humans, archea and many other organisms. Its principal function is to allow DNA replication to continue past DNA forks.
 W
WHomologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids. It is widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses.
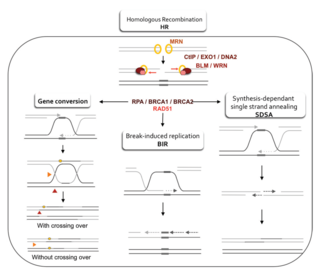 W
WHomology directed repair (HDR) is a mechanism in cells to repair double-strand DNA lesions. The most common form of HDR is homologous recombination. The HDR mechanism can only be used by the cell when there is a homologous piece of DNA present in the nucleus, mostly in G2 and S phase of the cell cycle. Other examples of homology-directed repair include single-strand annealing and breakage-induced replication. When the homologous DNA is absent, another process called non-homologous end joining (NHEJ) takes place instead.
 W
WIllegitimate recombination, or nonhomologous recombination, is the process by which two unrelated double stranded segments of DNA are joined. This insertion of genetic material which is not meant to be adjacent tends to lead to genes being broken causing the protein which they encode to not be properly expressed. One of the primary pathways by which this will occur is the repair mechanism known as non-homologous end joining (NHEJ).
 W
WKu is a dimeric protein complex that binds to DNA double-strand break ends and is required for the non-homologous end joining (NHEJ) pathway of DNA repair. Ku is evolutionarily conserved from bacteria to humans. The ancestral bacterial Ku is a homodimer. Eukaryotic Ku is a heterodimer of two polypeptides, Ku70 (XRCC6) and Ku80 (XRCC5), so named because the molecular weight of the human Ku proteins is around 70 kDa and 80 kDa. The two Ku subunits form a basket-shaped structure that threads onto the DNA end. Once bound, Ku can slide down the DNA strand, allowing more Ku molecules to thread onto the end. In higher eukaryotes, Ku forms a complex with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to form the full DNA-dependent protein kinase, DNA-PK. Ku is thought to function as a molecular scaffold to which other proteins involved in NHEJ can bind, orienting the double-strand break for ligation.
 W
WDNA ligase 4 is an enzyme that in humans is encoded by the LIG4 gene.
 W
WThe meiotic recombination checkpoint monitors meiotic recombination during meiosis, and blocks the entry into metaphase I if recombination is not efficiently processed.
 W
WThe microprocessor complex subunit DGCR8 (DiGeorge syndrome critical region 8) is a protein that in humans is encoded by the DGCR8 gene. In other animals, particularly the common model organisms Drosophila melanogaster and Caenorhabditis elegans, the protein is known as Pasha. It is a required component of the RNA interference pathway.
 W
WNon-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. NHEJ is referred to as "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair, which requires a homologous sequence to guide repair. The term "non-homologous end joining" was coined in 1996 by Moore and Haber.
 W
WNucleotide excision repair is a DNA repair mechanism. DNA damage occurs constantly because of chemicals, radiation and other mutagens. Three excision repair pathways exist to repair single stranded DNA damage: Nucleotide excision repair (NER), base excision repair (BER), and DNA mismatch repair (MMR). While the BER pathway can recognize specific non-bulky lesions in DNA, it can correct only damaged bases that are removed by specific glycosylases. Similarly, the MMR pathway only targets mismatched Watson-Crick base pairs.
 W
WO6-alkylguanine DNA alkyltransferase (also known as AGT, MGMT or AGAT) is a protein that in humans is encoded by the O6-methylguanine DNA methyltransferase (MGMT) gene. O6-methylguanine DNA methyltransferase is crucial for genome stability. It repairs the naturally occurring mutagenic DNA lesion O6-methylguanine back to guanine and prevents mismatch and errors during DNA replication and transcription. Accordingly, loss of MGMT increases the carcinogenic risk in mice after exposure to alkylating agents. The two bacterial isozymes are Ada and Ogt.
 W
W8-Oxoguanine (8-hydroxyguanine, 8-oxo-Gua, or OH8Gua) is one of the most common DNA lesions resulting from reactive oxygen species modifying guanine, and can result in a mismatched pairing with adenine resulting in G to T and C to A substitutions in the genome. In humans, it is primarily repaired by DNA glycosylase OGG1. It can be caused by ionizing radiation, in connection with oxidative metabolism.
 W
WPhotolyases are DNA repair enzymes that repair damage caused by exposure to ultraviolet light. These enzymes require visible light both for their own activation and for the actual DNA repair. The DNA repair mechanism involving photolyases is called photoreactivation. They mainly convert pyrimidine dimers into a normal pair of pyrimidine bases.
 W
WDNA polymerase kappa is an DNA polymerase that in humans is encoded by the POLK gene. It is involved in translesion synthesis.
 W
WPoly (ADP-ribose) polymerase (PARP) is a family of proteins involved in a number of cellular processes such as DNA repair, genomic stability, and programmed cell death.
 W
WProliferating cell nuclear antigen (PCNA) is a DNA clamp that acts as a processivity factor for DNA polymerase δ in eukaryotic cells and is essential for replication. PCNA is a homotrimer and achieves its processivity by encircling the DNA, where it acts as a scaffold to recruit proteins involved in DNA replication, DNA repair, chromatin remodeling and epigenetics.
 W
WE3 ubiquitin-protein ligase RAD18 is an enzyme that in humans is encoded by the RAD18 gene.
 W
WRAD51 is a eukaryotic gene. The enzyme encoded by this gene is a member of the RAD51 protein family which assists in repair of DNA double strand breaks. RAD51 family members are homologous to the bacterial RecA, Archaeal RadA and yeast Rad51. The protein is highly conserved in most eukaryotes, from yeast to humans.
 W
WRecA is a 38 kilodalton protein essential for the repair and maintenance of DNA. A RecA structural and functional homolog has been found in every species in which one has been seriously sought and serves as an archetype for this class of homologous DNA repair proteins. The homologous protein is called RAD51 in eukaryotes and RadA in archaea.
 W
WSLX4 is a protein involved in DNA repair, where it has important roles in the final steps of homologous recombination. Mutations in the gene are associated with the disease Fanconi anemia.
 W
WThe SOS response is a global response to DNA damage in which the cell cycle is arrested and DNA repair and mutagenesis is induced. The system involves the RecA protein. The RecA protein, stimulated by single-stranded DNA, is involved in the inactivation of the repressor (LexA) of SOS response genes thereby inducing the response. It is an error-prone repair system that contributes significantly to DNA changes observed in a wide range of species.
 W
WVery short patch (VSP) repair is a DNA repair system that removes GT mismatches created by the deamination of 5-methylcytosine to thymine. This system exists because the glycosylases which normally target deaminated bases cannot target thymine.
 W
WIn molecular biology, the XPG-I is a protein domain found on Xeroderma Pigmentosum Complementation Group G (XPG) protein. The XPG protein is an endonuclease which repairs DNA damage caused by ultraviolet light. The XPG protein repairs DNA by a process called, Nucleotide excision repair. Mutations in the protein commonly cause Xeroderma Pigmentosum which often lead to skin cancer.